The data standardization algorithm consists of two distinctive phases. Thus for each gene of a given expression matrix the following two steps will be performed:
A dedicated algorithm, called R-Radius Neighbours (RRN) algorithm, has been developed for the purpose of generating an estimation list for each gene profile. Such an estimation list consists of genes with expression profiles which exhibit at least minimum (preliminary determined) similarity in terms of DTW distance(1) to the expression profile of the gene in question. These profiles are consequently used for data standardization. The contribution of each gene in the estimation list is weighted by the degree of similarity in terms of DTW distance(2) of its expression profile to the expression profile of the gene under standardization.
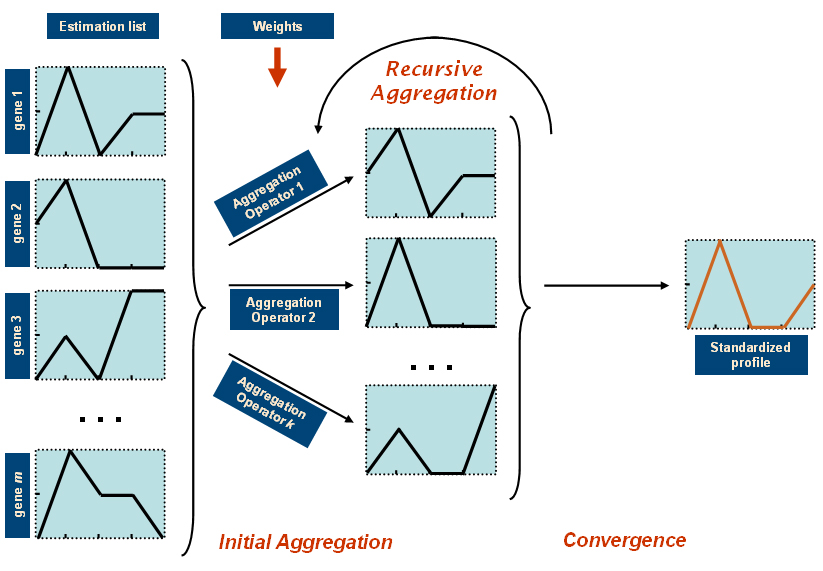
A recursive aggregation algorithm is applied to obtain the standardized expression profile of a gene by aggregating the expression profiles of the genes in its estimation list. The considered aggregation model has been inspired by a work on non-parametric recursive aggregation(3), where a set of aggregation operators is applied initially over a vector of input values, and then again over the result of the aggregation, and so on until a certain stop condition is met.
The used recursive aggregation algorith is illustrated in the below figure. The expression profiles included in the estimation list are initially combined in parallel with k different weighted aggregation operators. In this way k new expression profiles (one per aggregation operator) are produced and these new profiles are aggregated again this time with the nonparametric versions of the given aggregation operators. The latter process is repeated again and again until for each time point the difference between the aggregated values is small enough to stop further aggregation.
(1) Sakoe,H. and Chiba,S. (1978) Dynamic programming algorithm optimization for spoken word recognition, IEEE Trans. on Acoust., Speech, and Signal Process, ASSP-26, 43-49.
(2) Criel, J. and Tsiporkova, E. Gene Time Eχpression Warper: A tool for alignment, template matching and visualization of gene expression time series. Bioinformatics 22 2 (2006) 251-252.
(3) Tsiporkova, E. and Boeva, V. Nonparametric Recursive Aggregation Process. Kybernetika. J. of the Czech Society for Cybernetics and Inf. Sciences 40 1 (2004) 51-70. |
