A MULTI-PURPOSE TIME SERIES DATA
STANDARDIZATION APPROACH
Validation |
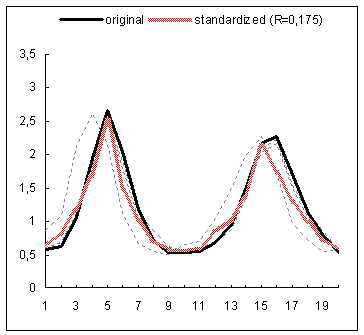
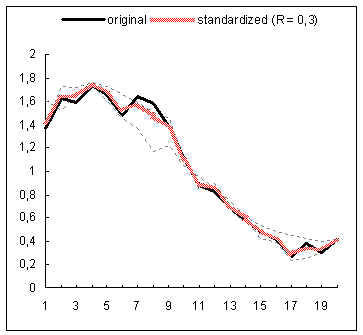
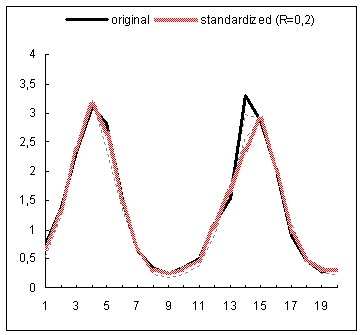
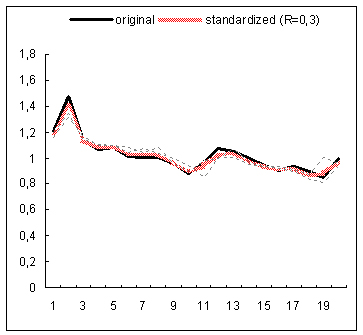
The proposed data standardization algorithm has been evaluated on gene expression time series data coming from a study examining the global cell-cycle control of gene expression in fission yeast Schizosaccharomyces pombe(1). The normalized data for 9 experiments has been downloaded from the website of the Sanger Institute. Subsequently, the rows with more than 25% missing entries have been filtered out from each expression matrix and any other missing expression entries have been imputed with the DTWimpute algorithm(2). In this way nine complete expression matrices have been obtained. Subsequently, the RRN based standardization method has been applied to each complete matrix. For each gene profile occurring in such a matrix a gene estimation list has been created by using the RRN algorithm. Thus for each gene profile a varying number of neighboring gene profiles has been identified and further used to calculate its standardized expression profile. The below figure depicts for 4 different genes the standardized and original expression profiles on the background of the profiles in the estimation list used for the standardization of each original profile. The standardized profiles depicted in the first and third plots, exhibit correction for their second peak shifts of the original profiles with respect to their neighboring profiles. The profile in the second plot is clearly smoothed by the standardization process. In fact it is an example of a clear fluctuation reduction as a result of the standardization procedure. The latter can easily be noticed in the upper and down parts of the standardized profile. In the fourth plot, the depicted standardized profile almost repeats the original one, which is obviously due to the closer match between the original profile and the profiles used for the standardization. In general, the presented results demonstrate that the standardization procedure operates as a sort of data correction for e.g., peak shifts, amplitude range, fluctuations, etc.
(1) Rustici,G., Mata, J., Kivinen, K., Lio, P., Penkett, C. J., Burns, G., Hayles, J., Brazma, A., Nurse, P., Bähler, J. (2004) Periodic gene expression program of the fission yeast cell cycle, Nature Genetics, 36, 809-817. (2) Tsiporkova,E. and Boeva,V. Two-pass imputation algorithm for missing value estimation in gene expression time series. Journal of Bioinformatics and Computational Biology, 5 5 (October 2007) 1005-1022. |